![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | WT14_S1_R1_001.fastq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 1038333 |
| Filtered Sequences | 0 |
| Sequence length | 151 |
| %GC | 41 |
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality

![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores

![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content

![[WARN]](Icons/warning.png) Per base GC content
Per base GC content

![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content

![[OK]](Icons/tick.png) Per base N content
Per base N content

![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution

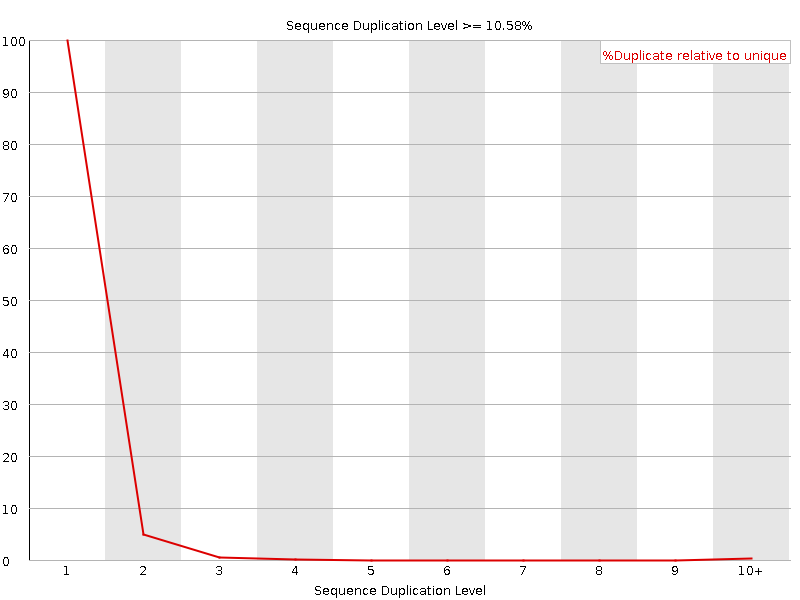
![[OK]](Icons/tick.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACAGTCAACAATCTCGTAT | 1713 | 0.16497597591524107 | TruSeq Adapter, Index 8 (97% over 36bp) |
| GATCTGGTTTTATTACATTCTGAATTGGACGTTGAAAATGAGCTTATCTC | 1656 | 0.15948640753977772 | No Hit |
| GATCCAGCCATAAAATGCATCATTCTTTTTTGTTTTAGACAACATTTCAT | 1411 | 0.13589089434699658 | No Hit |
| GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG | 1151 | 0.11085075789751457 | No Hit |
| GATCCAGCTATAAAATGCATCATTCTTTTTTGTTTTAGACAACATTTCAT | 1144 | 0.11017660037772083 | No Hit |
![[FAIL]](Icons/error.png) Kmer Content
Kmer Content
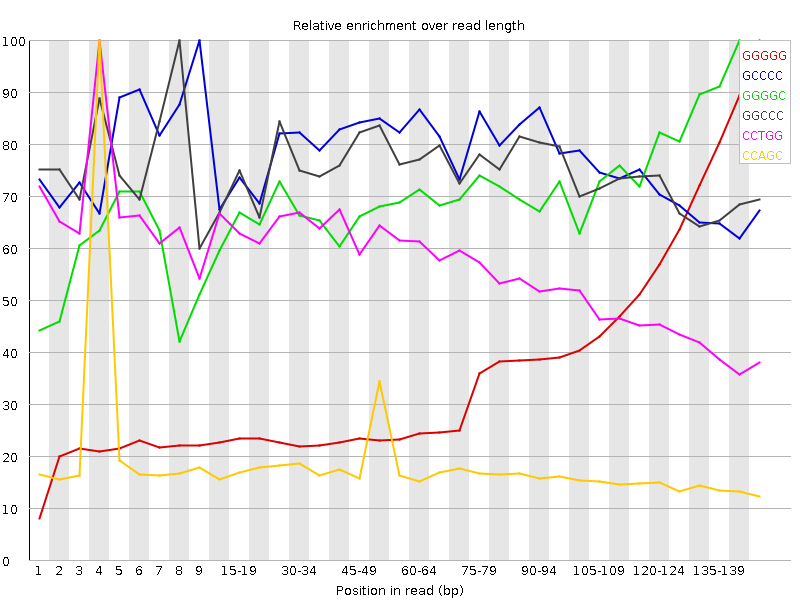
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GGGGG | 947525 | 13.914402 | 36.00961 | 145-147 |
| GCCCC | 180650 | 3.2882028 | 4.266316 | 9 |
| GGGGC | 201295 | 3.1190228 | 4.3479047 | 140-144 |
| GGCCC | 177725 | 3.0658963 | 4.1065073 | 8 |
| CCTGG | 227845 | 2.7770457 | 5.0070844 | 4 |
| CCAGC | 207760 | 2.5803185 | 15.005232 | 4 |
| GATCG | 301045 | 2.5036044 | 11.134491 | 1 |
| TCCAG | 246680 | 2.164611 | 12.400946 | 3 |
| CAGCC | 153620 | 1.9079155 | 9.410215 | 5 |
| TTTTA | 551555 | 1.8011519 | 5.561522 | 8 |
| GTTTT | 377685 | 1.713124 | 5.8224 | 7 |
| GATCT | 268040 | 1.6618054 | 28.309978 | 1 |
| GATCC | 180190 | 1.581163 | 30.957466 | 1 |
| TTTAT | 461330 | 1.5065142 | 5.012353 | 9 |
| GATCA | 249375 | 1.4931024 | 24.91118 | 1 |
| ATCTG | 233370 | 1.446857 | 13.681579 | 2 |
| AGCCA | 160955 | 1.3639756 | 6.651149 | 6 |
| TCTGG | 154745 | 1.3325845 | 9.227099 | 3 |
| ATCAG | 213965 | 1.2810895 | 8.218904 | 2 |
| ATCCA | 201425 | 1.2725118 | 12.938552 | 2 |
| ATCTC | 181355 | 1.1863745 | 6.4176173 | 2 |
| CTGGT | 134395 | 1.1573409 | 7.8917656 | 4 |
| ATCAT | 254880 | 1.1376766 | 5.576122 | 2 |
| ATCAC | 179265 | 1.132515 | 5.4270425 | 2 |
| GGTTT | 183040 | 1.1136757 | 6.7382836 | 6 |
| ATCGG | 125460 | 1.043373 | 5.060168 | 2 |
| TGGTT | 169115 | 1.0289514 | 5.517692 | 5 |
| ATCAA | 230900 | 0.995321 | 5.866519 | 2 |
| ATCCC | 100250 | 0.928201 | 6.1439223 | 2 |
| ATCCT | 131115 | 0.8577182 | 5.3456106 | 2 |