![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | SOX18_S5_R1_001.fastq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 754700 |
| Filtered Sequences | 0 |
| Sequence length | 151 |
| %GC | 42 |
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality

![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores

![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content

![[WARN]](Icons/warning.png) Per base GC content
Per base GC content

![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content

![[OK]](Icons/tick.png) Per base N content
Per base N content

![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution

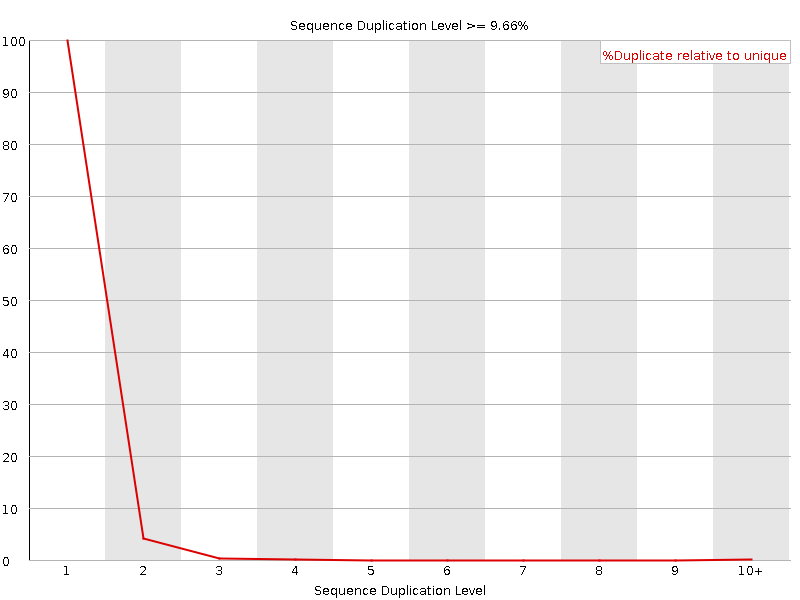
![[OK]](Icons/tick.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG | 1450 | 0.19212932290976545 | No Hit |
| GATCTGGTTTTATTACATTCTGAATTGGACGTTGAAAATGAGCTTATCTC | 1269 | 0.16814628329137404 | No Hit |
| GATCCAGCCATAAAATGCATCATTCTTTTTTGTTTTAGACAACATTTCAT | 1177 | 0.15595600901020273 | No Hit |
| GATCCAGCTATAAAATGCATCATTCTTTTTTGTTTTAGACAACATTTCAT | 1118 | 0.14813833311249502 | No Hit |
![[FAIL]](Icons/error.png) Kmer Content
Kmer Content
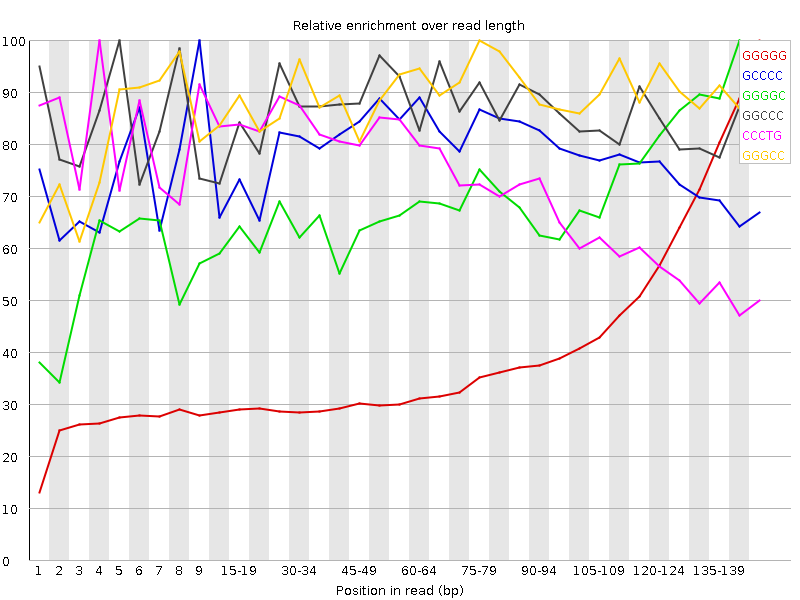
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GGGGG | 716900 | 13.367759 | 32.134014 | 145-147 |
| GCCCC | 143395 | 3.3545196 | 4.297267 | 9 |
| GGGGC | 159650 | 3.1505983 | 4.4899774 | 140-144 |
| GGCCC | 141415 | 3.125847 | 3.6388369 | 5 |
| CCCTG | 182590 | 3.076215 | 4.296524 | 4 |
| GGGCC | 144555 | 3.0191267 | 3.3701587 | 75-79 |
| CCTGG | 181970 | 2.8967793 | 5.054142 | 4 |
| CCAGC | 159380 | 2.6158383 | 16.332167 | 4 |
| TCCAG | 178965 | 2.1153803 | 13.916438 | 3 |
| GATCG | 172690 | 1.928695 | 11.678394 | 1 |
| CAGCC | 114655 | 1.8817853 | 9.721973 | 5 |
| TTTTA | 391675 | 1.8302077 | 5.7883344 | 8 |
| CAGCT | 146890 | 1.7362516 | 5.4292865 | 5 |
| GTTTT | 272920 | 1.7175332 | 5.8307385 | 7 |
| GATCT | 192290 | 1.6368937 | 32.50861 | 1 |
| TTTAT | 331405 | 1.5485798 | 5.658323 | 9 |
| GATCC | 130860 | 1.5467757 | 35.541676 | 1 |
| ATCTG | 167860 | 1.4289299 | 15.139243 | 2 |
| AGCCA | 117245 | 1.350057 | 7.0322666 | 6 |
| GATCA | 160175 | 1.328299 | 25.593655 | 1 |
| TCTGG | 114960 | 1.3179706 | 9.209251 | 3 |
| ATCCA | 146845 | 1.2887964 | 14.918523 | 2 |
| ATCAG | 148495 | 1.2314389 | 7.392425 | 2 |
| ATCTC | 128165 | 1.1546683 | 7.819208 | 2 |
| CTGGT | 100135 | 1.1480079 | 7.6758313 | 4 |
| ATCAC | 127850 | 1.1220853 | 5.8435717 | 2 |
| GGTTT | 134280 | 1.108699 | 6.60795 | 6 |
| ATCAT | 175400 | 1.1086594 | 5.6159067 | 2 |
| TGGTT | 124990 | 1.0319949 | 5.54614 | 5 |
| ATCAA | 162965 | 1.0034604 | 6.7288666 | 2 |
| ATCTT | 139910 | 0.9077785 | 5.8601413 | 2 |
| ATCCC | 71715 | 0.89712816 | 6.65592 | 2 |
| ATCCT | 95800 | 0.86308455 | 6.197103 | 2 |