![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | NAN21-22_S4_R2_001.fastq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 294324 |
| Filtered Sequences | 0 |
| Sequence length | 151 |
| %GC | 41 |
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality

![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores

![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content

![[WARN]](Icons/warning.png) Per base GC content
Per base GC content

![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content

![[OK]](Icons/tick.png) Per base N content
Per base N content

![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution

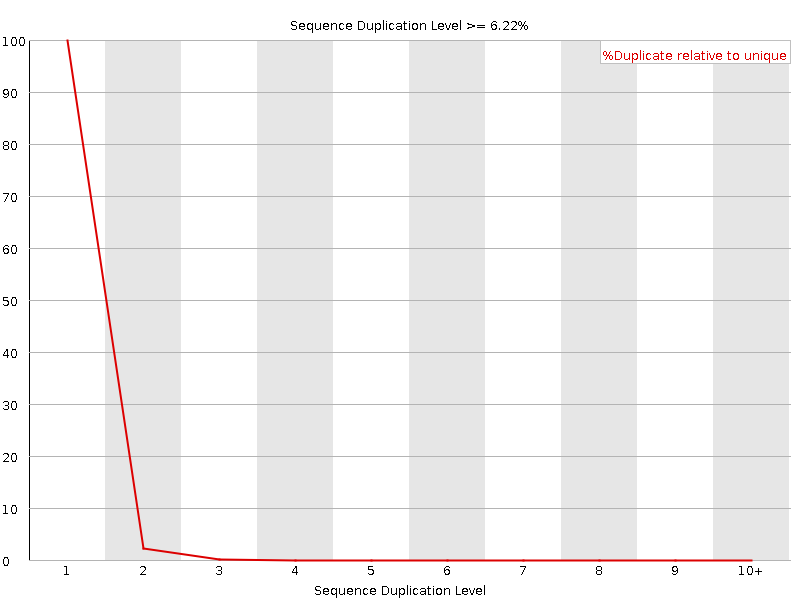
![[OK]](Icons/tick.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GATCTGGTTTTATTACATTCTGAATTGGACGTTGAAAATGAGCTTATCTC | 346 | 0.1175575216428154 | No Hit |
| GATCCAGCCATAAAATGCATCATTCTTTTTTGTTTTAGACAACATTTCAT | 341 | 0.11585871352658975 | No Hit |
![[FAIL]](Icons/error.png) Kmer Content
Kmer Content
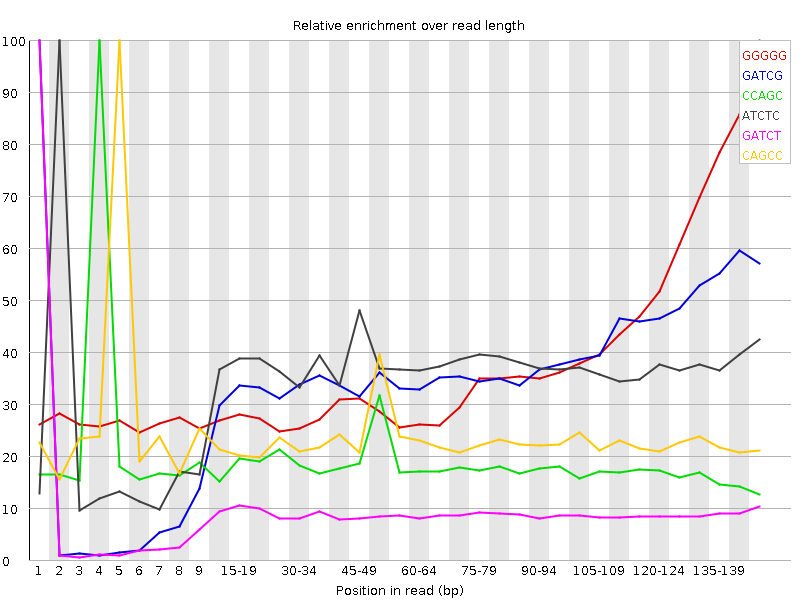
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GGGGG | 205040 | 10.998021 | 27.921062 | 145-147 |
| GATCG | 99435 | 2.9467678 | 7.8091803 | 1 |
| CCAGC | 57600 | 2.5468855 | 13.897175 | 4 |
| ATCTC | 89210 | 2.0476377 | 5.5699043 | 2 |
| GATCT | 92515 | 2.0235503 | 22.249777 | 1 |
| CAGCC | 43555 | 1.9258611 | 8.279859 | 5 |
| TCCAG | 61510 | 1.9128904 | 11.21929 | 3 |
| GATCA | 71160 | 1.5043201 | 19.279308 | 1 |
| ATCTG | 63135 | 1.3809311 | 11.065843 | 2 |
| AGCCA | 44985 | 1.3521183 | 5.9387956 | 6 |
| TCTGG | 43645 | 1.3382542 | 7.8626523 | 3 |
| ATCCA | 55875 | 1.2395375 | 11.125528 | 2 |
| CTGGT | 37870 | 1.1611798 | 6.2820897 | 4 |
| GGTTT | 53085 | 1.14481 | 5.2747874 | 6 |
| ATCAG | 53885 | 1.1391273 | 6.2025666 | 2 |
| GATCC | 35920 | 1.1170709 | 24.378658 | 1 |
| ATCAA | 60750 | 0.9161115 | 5.1009855 | 2 |