![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | NAN21-22_S4_R1_001.fastq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 294324 |
| Filtered Sequences | 0 |
| Sequence length | 151 |
| %GC | 41 |
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality

![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores

![[WARN]](Icons/warning.png) Per base sequence content
Per base sequence content

![[WARN]](Icons/warning.png) Per base GC content
Per base GC content

![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content

![[OK]](Icons/tick.png) Per base N content
Per base N content

![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution

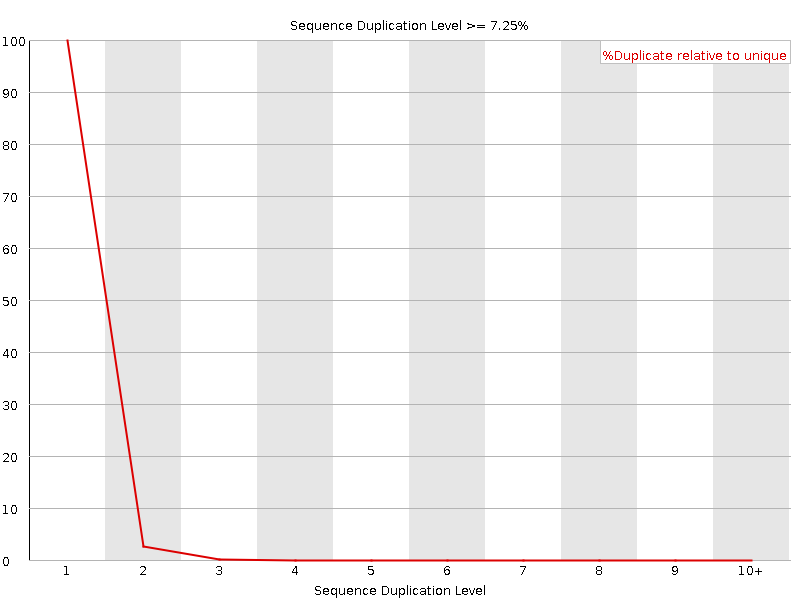
![[OK]](Icons/tick.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG | 522 | 0.17735556733395852 | No Hit |
| GATCTGGTTTTATTACATTCTGAATTGGACGTTGAAAATGAGCTTATCTC | 350 | 0.11891656813579593 | No Hit |
| TGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG | 333 | 0.11314062054062869 | No Hit |
| GATCCAGCCATAAAATGCATCATTCTTTTTTGTTTTAGACAACATTTCAT | 314 | 0.1066851496989712 | No Hit |
![[FAIL]](Icons/error.png) Kmer Content
Kmer Content
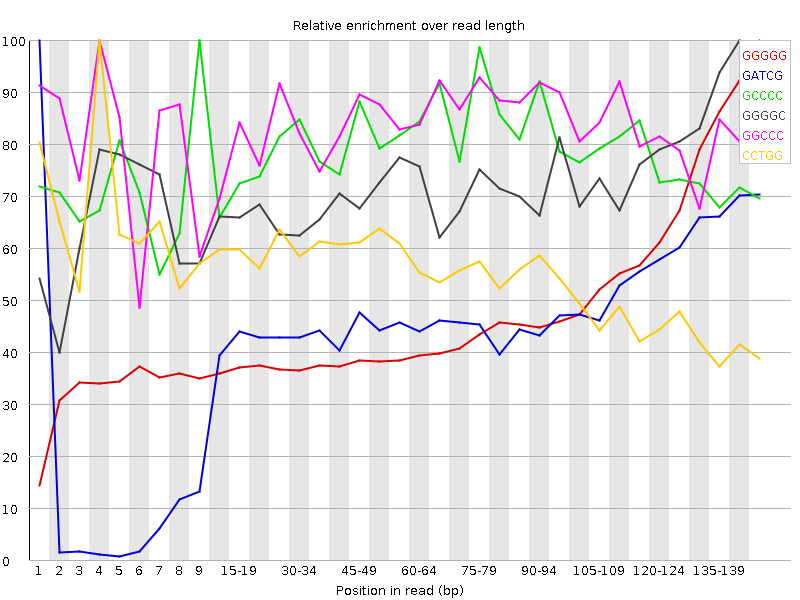
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GGGGG | 258235 | 13.3426485 | 27.289558 | 145-147 |
| GATCG | 111835 | 3.2728577 | 6.946312 | 1 |
| GCCCC | 51625 | 3.2360926 | 4.098945 | 9 |
| GGGGC | 56200 | 3.047521 | 4.17873 | 140-144 |
| GGCCC | 50555 | 3.0195446 | 3.5998313 | 4 |
| CCTGG | 65295 | 2.767748 | 5.1095357 | 4 |
| CCAGC | 59305 | 2.56658 | 11.069646 | 4 |
| TCCAG | 69935 | 2.1479661 | 9.278083 | 3 |
| CAGCC | 43080 | 1.8644005 | 6.488441 | 5 |
| GATCC | 54845 | 1.6844956 | 22.028505 | 1 |
| GATCT | 74055 | 1.6141968 | 21.736237 | 1 |
| ATCTG | 65635 | 1.4306638 | 11.229665 | 2 |
| TCTGG | 44595 | 1.3415353 | 6.9648232 | 3 |
| ATCCA | 59530 | 1.324814 | 9.322675 | 2 |
| GATCA | 60430 | 1.2814101 | 18.68345 | 1 |
| ATCAG | 56470 | 1.1974388 | 5.984301 | 2 |
| CTGGT | 37950 | 1.1416361 | 5.5498414 | 4 |
| ATCAA | 64890 | 0.9970099 | 5.2620416 | 2 |