![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | NAN20_S3_R2_001.fastq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 1020808 |
| Filtered Sequences | 0 |
| Sequence length | 151 |
| %GC | 41 |
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality

![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores

![[WARN]](Icons/warning.png) Per base sequence content
Per base sequence content

![[WARN]](Icons/warning.png) Per base GC content
Per base GC content

![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content

![[OK]](Icons/tick.png) Per base N content
Per base N content

![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution

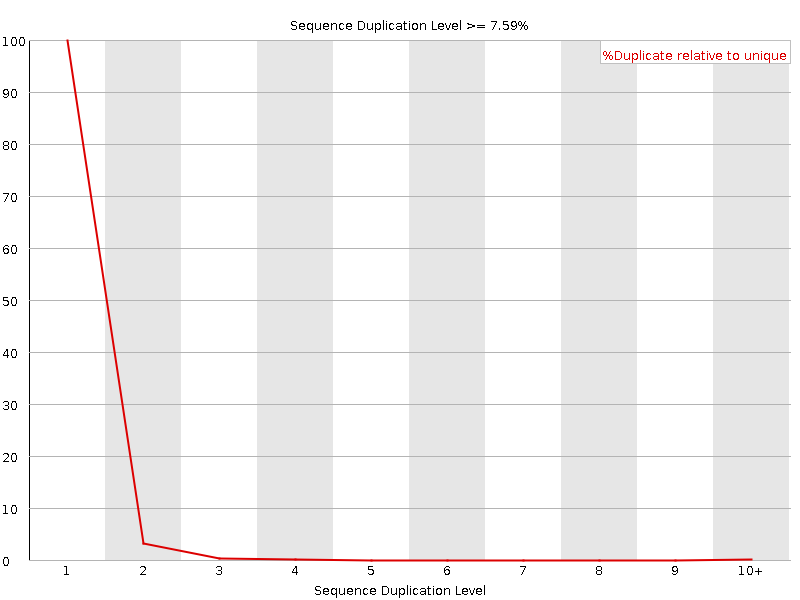
![[OK]](Icons/tick.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG | 1328 | 0.13009302434933895 | No Hit |
| GATCTGGTTTTATTACATTCTGAATTGGACGTTGAAAATGAGCTTATCTC | 1096 | 0.107365929734093 | No Hit |
![[FAIL]](Icons/error.png) Kmer Content
Kmer Content
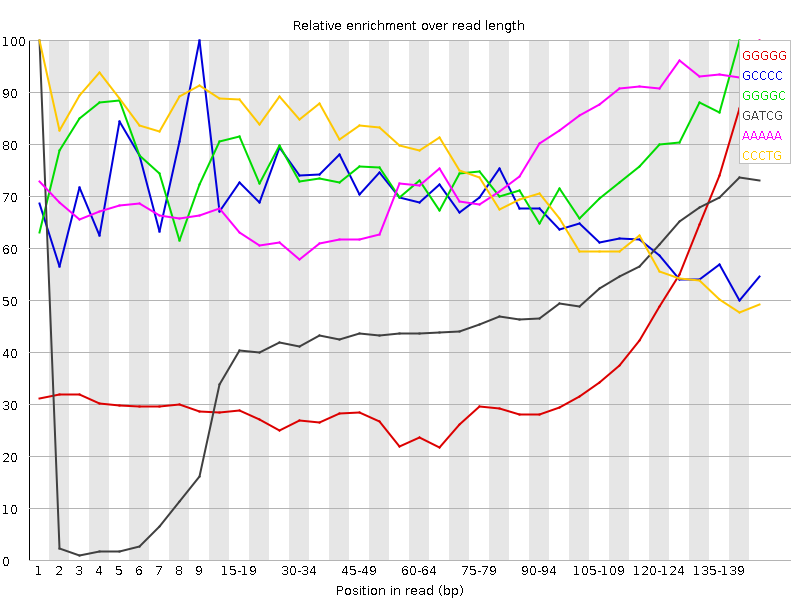
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GGGGG | 847905 | 12.516361 | 34.188633 | 145-147 |
| GCCCC | 172465 | 3.2935858 | 4.9074664 | 9 |
| GGGGC | 207390 | 3.2649608 | 4.2967196 | 140-144 |
| GATCG | 382935 | 3.2407503 | 6.783674 | 1 |
| AAAAA | 1094085 | 3.0982485 | 4.087199 | 145-147 |
| CCCTG | 230145 | 3.0821497 | 4.258459 | 1 |
| GGCCC | 170600 | 3.0548346 | 4.1549315 | 8 |
| CCAGC | 198510 | 2.5549242 | 11.60642 | 4 |
| TCCAG | 216740 | 1.9562253 | 9.630263 | 3 |
| CAGCC | 143475 | 1.8465956 | 7.144164 | 5 |
| GATCT | 287360 | 1.8188223 | 19.241129 | 1 |
| GATCA | 258885 | 1.5747596 | 18.522919 | 1 |
| ATCTG | 215775 | 1.3657308 | 9.626123 | 2 |
| AGCCA | 152525 | 1.3230132 | 5.0577145 | 6 |
| TCTGG | 149495 | 1.3164482 | 6.3738976 | 3 |
| ATCCA | 195040 | 1.2652909 | 9.775648 | 2 |
| GATCC | 134250 | 1.2116971 | 21.485048 | 1 |
| ATCAG | 185285 | 1.1270616 | 6.4194264 | 2 |
| CTGGT | 127370 | 1.1216161 | 5.5419455 | 4 |