![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | NAN20_S3_R1_001.fastq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 1020808 |
| Filtered Sequences | 0 |
| Sequence length | 151 |
| %GC | 41 |
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality

![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores

![[WARN]](Icons/warning.png) Per base sequence content
Per base sequence content

![[WARN]](Icons/warning.png) Per base GC content
Per base GC content

![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content

![[OK]](Icons/tick.png) Per base N content
Per base N content

![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution

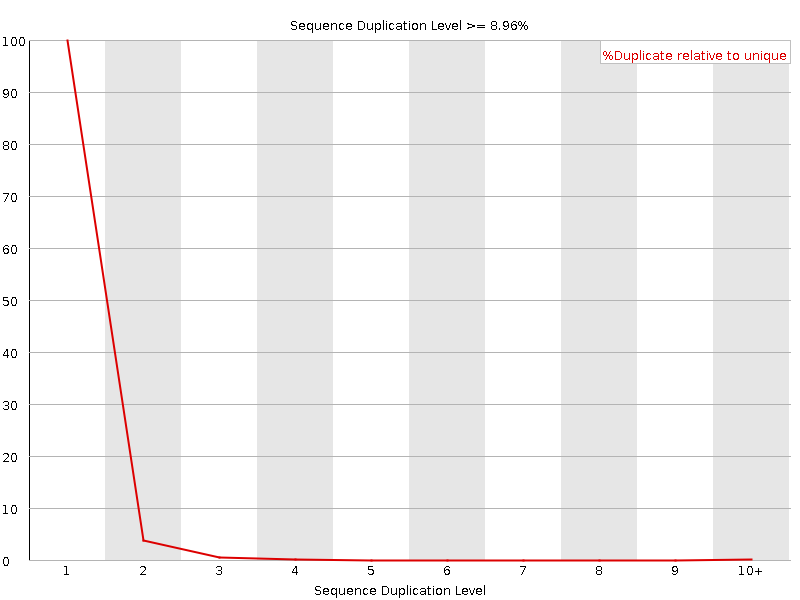
![[OK]](Icons/tick.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG | 1847 | 0.1809351023894797 | No Hit |
| TGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG | 1477 | 0.14468930494275123 | No Hit |
| GATCTGGTTTTATTACATTCTGAATTGGACGTTGAAAATGAGCTTATCTC | 1070 | 0.1048189277513499 | No Hit |
![[FAIL]](Icons/error.png) Kmer Content
Kmer Content
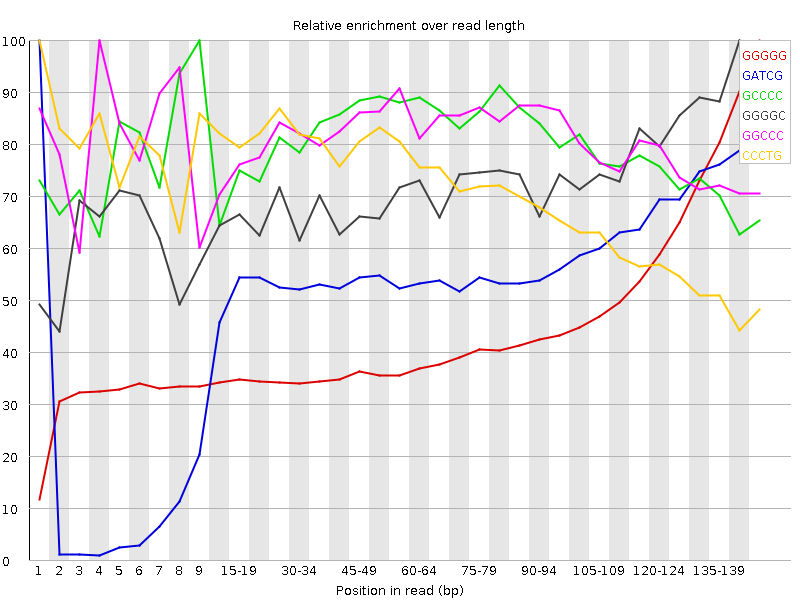
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GGGGG | 1049670 | 15.031044 | 32.73486 | 145-147 |
| GATCG | 425950 | 3.5611732 | 6.3403797 | 1 |
| GCCCC | 189710 | 3.4395192 | 4.3161464 | 9 |
| GGGGC | 214485 | 3.258 | 4.450054 | 140-144 |
| GGCCC | 187995 | 3.213187 | 3.9818816 | 4 |
| CCCTG | 245040 | 3.1514792 | 4.498684 | 1 |
| GGGCC | 192720 | 3.1052651 | 3.3652709 | 60-64 |
| CCAGG | 255110 | 3.0067134 | 3.6802917 | 8 |
| CCAGC | 206310 | 2.5793045 | 10.814294 | 4 |
| TCCAG | 246485 | 2.1859627 | 9.059679 | 3 |
| CAGCC | 147015 | 1.8379933 | 6.964515 | 5 |
| GATCC | 194440 | 1.7243994 | 20.646122 | 1 |
| GATCT | 250315 | 1.5747408 | 18.297808 | 1 |
| ATCTG | 226795 | 1.4267756 | 9.389806 | 2 |
| ATCCA | 207305 | 1.3447906 | 9.172219 | 2 |
| TCTGG | 152780 | 1.3140023 | 6.0680146 | 3 |
| GATCA | 213215 | 1.3039017 | 17.872437 | 1 |
| ATCAG | 197020 | 1.2048622 | 6.264903 | 2 |